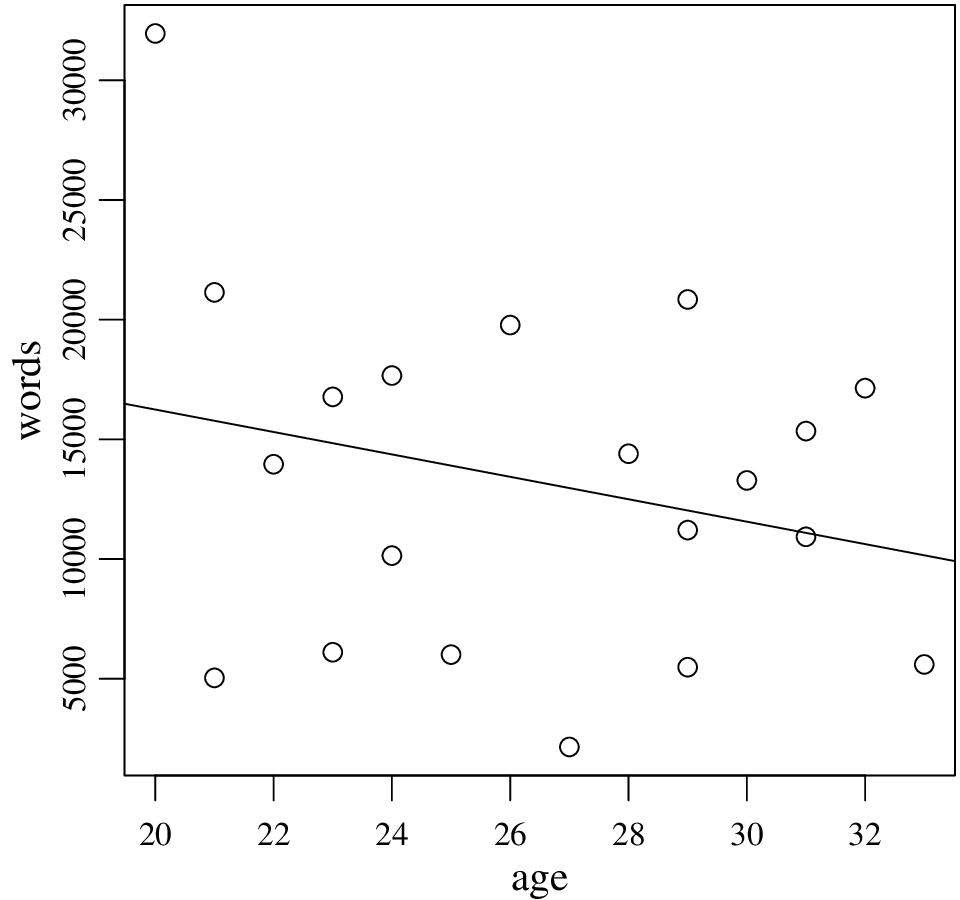
Answer of 2.8. Here is the result, including the scatter plot:
The regression line indeed agrees, it indicates that as age increases, people become less talkative. In fact, the correlation coefficient, Pearson’s r, is simply normalized simple regression slope. However, as we will stress later, least squares regression (and also correlation) is sensitive to extreme values. In our example, it seems the regression line is affected (possibly substantially) by the youngest and most talkative participant.
You may want to remove this data point (or replace with a more moderate value) and try last three exercises again. You are encoraged to exercise with removing or replacing data points as you like here. However, when you remove data points in ‘real’ research, you should be convinced (and be ready to convince others) that it is a sensible thing to do.